Science Projects

THE PROJECT
Science Projects
The project and the Lab will be focusing on two main themes. First, the advancement of technology towards enabling the investigation of challenging protein samples and secondly the exploitation of such technology to unravel disease-related states.

Science Projects
A. Technology Development
Politis Lab has developed HDX-MS strategies to interrogate membrane proteins, including transporters and G-Protein Coupled Receptors (GPCRs), within their native lipid environment where they live and operate. We have demonstrated that HDX-MS can be used as a drug profiling tool to monitor ligand binding to GPCRs, and sort them into different categories based on the allosteric effects observed. In addition to our leading place in the field of research, we have made significant progress in HDX-MS of IMPs in its native environment. This remains a challenge due to the high quantity of lipids that can induce peptide ionisation suppression and impair chromatographic separation of peptides. Here we aim to develop in situ HDX-MS for interrogating the conformational dynamics of membrane proteins in their native environment.

Technology development
Liposomes
Reconstitution of IMPs into liposomes offers all the benefits of other mimetics (e.g. fine-tuning lipid composition) with the addition of curvature, lateral pressure and protein directionality that mimic biological membranes. Liposomes are used to study multicomponent systems due to their large size, and their aqueous lumen not only allows for the maintenance of membrane potential, but can also encapsulate activity reporter systems. Despite advantages, the use of liposomes in MS remains limited to small peptides or single-pass transmembrane proteins, owing to the difficulty in handling and analysing heterogeneity of samples in current systems. In our group building upon advances in handling heterogeneous samples, we generate single protein orientation liposomes with high levels of reproducibility. Additionally, we utilise the positive-inside rule to correctly insert protein into native membranes. This unique directionality not only allows for the application of energy gradients, but it also reduces the limitation of falsely identified kinetics in HDX-MS, offering high quality data.

Technology development
Native membranes
While liposomes provide the best simplistic mimetic, isolated membrane vesicles provide the best option to study MPs in native lipid bilayers. Isolated membrane vesicles have the added advantage of maintaining membrane associated protein interaction partners, as well as electrochemical gradient housekeepers. These membranes should also have more lipid asymmetry, which is challenging to achieve with synthetic vesicles. We utilise our delipidation strategy to fingerprint overexpressed proteins from E. coli cells, followed by harvesting the inner membranes and subsequently solubilising them in lipid vesicles. We analyse interactions in situ by carrying out HDX-MS in native membranes.

Science Projects
B. Applications
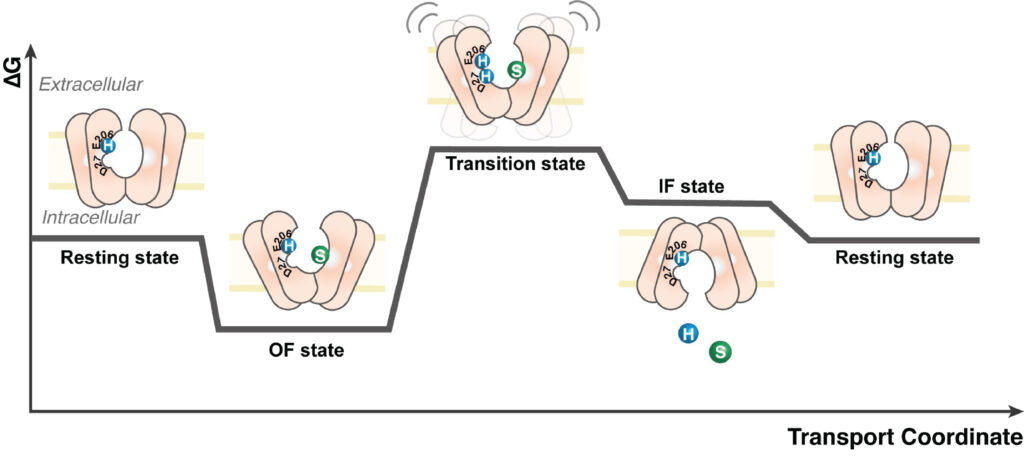
Transporters
Transporters are essential biomolecules responsible for the movement of substrates across biological membranes. Structural information about the conformations adopted by transporters remains scarce primarily due to the challenges faced by traditional structural biology approaches. The challenge is even greater for eukaryotic systems due to their intrinsic dynamics and the low yield of purification. To overcome these, we employ HDX-MS to capture the conformational states of transporters including sugar transporters (XylE, LacY) and its human homologue, GLUT1. We have established mechanistic details on how the lipid environment affects the conformational dynamics of sugar transporters. This has led to a comprehensive HDX-MS protocol for carrying out experiments in lipid nanodiscs and combining them with molecular dynamics simulations. We have shown how ligand binding impacts structural dynamics and also establish the role of lipids on oligomeric states of eukaryotic transporters
- Martens, C., Shekhar, M., Lau, A.M., Tajkhorshid, E., and Politis, A. (2019). Integrating hydrogen-deuterium exchange mass spectrometry with molecular dynamics simulations to probe lipid-modulated conformational changes in membrane proteins. Nat Protoc 14, 3183-3204. 10.1038/s41596-019-0219-6.
- Pyle, E., Kalli, A.C., Amillis, S., Hall, Z., Lau, A.M., Hanyaloglu, A.C., Diallinas, G., Byrne, B., and Politis, A. (2018). Structural Lipids Enable the Formation of Functional Oligomers of the Eukaryotic Purine Symporter UapA. Cell Chem Biol 25, 840-848 e844. 10.1016/j.chembiol.2018.03.011..
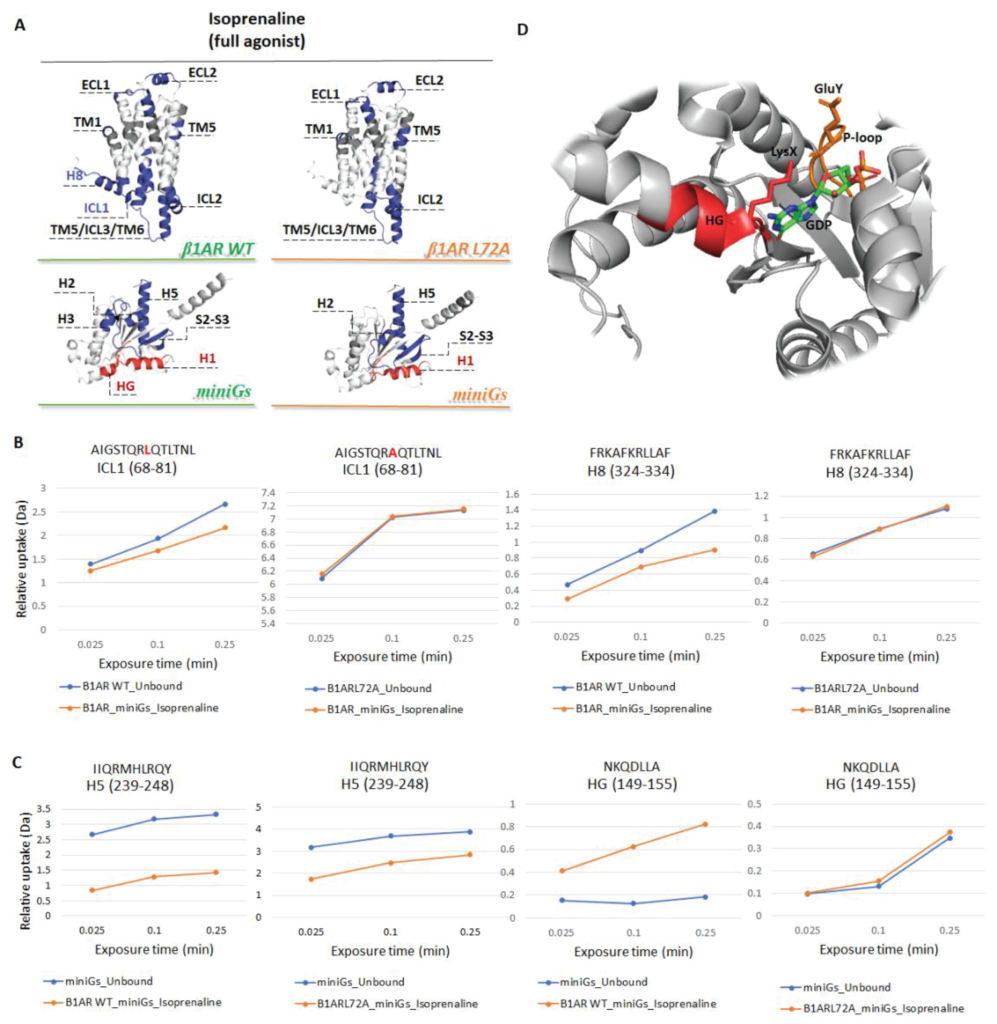
GPCRs
Probing the conformational dynamics of GPCRs in native environment
Collaboration with Dr G. Skretas, BSRC Fleming
HDX-MS provides a rather sensitive tool to interrogate the structural dynamics of membrane proteins by monitoring the exchange of hydrogen to bulk deuterium at native temperature. Our group has recently developed strategies to monitor structural changes of eukaryotic proteins including G protein-coupled receptors (GPCRs). We employed HDX-MS to characterise the structural dynamics of β1-adrenergic receptor (tβ1AR) in a complex with nine ligands, including agonists, partial agonists and antagonists3. We show that dynamic signatures across the GPCR structure can be grouped by compound modality. Surprisingly, we discovered repeated destabilisation of the intracellular loop 1 (ICL1) upon full agonist binding and stabilisation upon antagonist binding, suggesting that increased dynamics in this region are an essential component for G-protein recruitment.
We collaborate with Skretas Lab to interrogate the conformational dynamics of membrane proteins in their native environment. The Skretas Lab has recently generated the first set of specialized low-toxicity E. coli strains for enhanced production of recombinant MPs, termed SuptoxD and SuptoxR, and their derivatives6. This results in dramatically enhanced volumetric yields of well-folded recombinant MPs of both prokaryotic and eukaryotic origin, including the neurotensin receptor 1 (NTSR1). We utilise HDX-MS, which offers a highly sensitive tool to interrogate the structural dynamics of NTSR1 – a GPCR implicated in a wide range of physiological processes including blood pressure, analgesia and hypothermia – by monitoring the exchange of hydrogen to bulk deuterium. We investigate how PIP2 lipid binding impacts on structural dynamics and results in allosteric changes in NTSR1. We further explore the modulatory effect of lipids in eukaryotic proteins using the specialised E. coli strains, and study membrane proteins in situ by solubilising the MPs in lipid vesicles directly extracted from inner membranes. Our approach will not only establish in situ HDX-MS, but it will also transform the technique into a quantitative tool for the analysis of complex biological systems.
- Toporowska J,Kapoor P, Musgarrad M, Gherbi K, Sengmany K, Qu F, Yen HY, Hansen K, Jazayeri A, Hopper JTS, Politis A. Ligand-induced conformational changes in the β1-adrenergic receptor revealed by hydrogen deuterium exchange mass spectrometry. Nature Communications, 15, 8993, 2024, doi:10.1038/s41467-024-53161-0
- Michou M, Kapsalis C, Pliotas C, Skretas G. Optimization of recombinant membrane protein production in the engineered Escherichia coli strains SuptoxD and SuptoxR. ACS Synthetic Biology, 8(7), 1631-41, 2019. doi.101021/acssynbio.9b00120